Note
Click here to download the full example code
Compare PAC of two experimental conditions with cluster-based statistics¶
This example illustrates how to statistically compare the phase-amplitude coupling results coming from two experimental conditions. In particular, the script below a the cluster-based approach to correct for the multiple comparisons.
In order to work, this script requires MNE-Python package to be installed in
order to perform the cluster-based correction
(mne.stats.permutation_cluster_test())
import numpy as np
from tensorpac import Pac
from tensorpac.signals import pac_signals_wavelet
from mne.stats import permutation_cluster_test
import matplotlib.pyplot as plt
Simulate the data coming from two experimental conditions¶
Let’s start by simulating data coming from two experimental conditions. The first dataset is going to simulate a 10hz phase <-> 120hz amplitude coupling while the second dataset will not include any coupling (random data)
# create the first dataset with a 10hz <-> 100hz coupling
n_epochs = 30 # number of datasets
sf = 512. # sampling frequency
n_times = 4000 # Number of time points
x_1, time = pac_signals_wavelet(sf=sf, f_pha=10, f_amp=120, noise=2.,
n_epochs=n_epochs, n_times=n_times)
# create a second random dataset without any coupling
x_2 = np.random.rand(n_epochs, n_times)
Compute the single trial PAC on both datasets¶
once the datasets created, we can now extract the PAC, computed across time-points for each trials and across several phase and amplitude frequencies
# create the pac object. Use the Gaussian-Copula PAC
p = Pac(idpac=(6, 0, 0), f_pha='hres', f_amp='hres', dcomplex='wavelet')
# compute pac for both dataset
pac_1 = p.filterfit(sf, x_1, n_jobs=-1)
pac_2 = p.filterfit(sf, x_2, n_jobs=-1)
Correct for multiple-comparisons using a cluster-based approach¶
Then, we perform the cluster-based correction for multiple comparisons between the PAC coming from the two conditions. To this end we use the Python package MNE-Python and in particular, the function
mne.stats.permutation_cluster_test()
# mne requires that the first is represented by the number of trials (n_epochs)
# Therefore, we transpose the output PACs of both conditions
pac_r1 = np.transpose(pac_1, (2, 0, 1))
pac_r2 = np.transpose(pac_2, (2, 0, 1))
n_perm = 1000 # number of permutations
tail = 1 # only inspect the upper tail of the distribution
# perform the correction
t_obs, clusters, cluster_p_values, h0 = permutation_cluster_test(
[pac_r1, pac_r2], n_permutations=n_perm, tail=tail)
Out:
Using a threshold of 4.006873
/home/circleci/project/examples/stats/plot_compare_cond_cluster.py:71: DeprecationWarning: The default for "out_type" will change from "mask" to "indices" in version 0.22. To avoid this warning, explicitly set "out_type" to one of its string values.
[pac_r1, pac_r2], n_permutations=n_perm, tail=tail)
stat_fun(H1): min=0.000000 max=52.333180
Running initial clustering
Found 13 clusters
Permuting 999 times...
0%| | : 0/999 [00:00<?, ?it/s]
3%|2 | : 25/999 [00:00<00:01, 710.88it/s]
5%|5 | : 53/999 [00:00<00:01, 715.89it/s]
8%|7 | : 77/999 [00:00<00:01, 715.62it/s]
11%|# | : 106/999 [00:00<00:01, 721.62it/s]
14%|#3 | : 135/999 [00:00<00:01, 727.40it/s]
16%|#5 | : 158/999 [00:00<00:01, 724.90it/s]
19%|#8 | : 187/999 [00:00<00:01, 730.49it/s]
22%|##1 | : 215/999 [00:00<00:01, 734.75it/s]
24%|##4 | : 241/999 [00:00<00:01, 736.31it/s]
27%|##6 | : 268/999 [00:00<00:00, 739.12it/s]
29%|##9 | : 291/999 [00:00<00:00, 735.92it/s]
31%|###1 | : 311/999 [00:00<00:00, 727.03it/s]
33%|###3 | : 330/999 [00:00<00:00, 716.55it/s]
35%|###5 | : 353/999 [00:00<00:00, 714.59it/s]
38%|###8 | : 380/999 [00:00<00:00, 718.31it/s]
41%|####1 | : 411/999 [00:00<00:00, 726.10it/s]
44%|####3 | : 435/999 [00:00<00:00, 725.21it/s]
46%|####6 | : 461/999 [00:00<00:00, 727.15it/s]
49%|####9 | : 493/999 [00:00<00:00, 735.62it/s]
51%|#####1 | : 512/999 [00:00<00:00, 724.28it/s]
53%|#####3 | : 530/999 [00:00<00:00, 711.37it/s]
55%|#####4 | : 549/999 [00:00<00:00, 702.08it/s]
57%|#####6 | : 566/999 [00:00<00:00, 688.17it/s]
59%|#####8 | : 587/999 [00:00<00:00, 684.39it/s]
61%|###### | : 608/999 [00:00<00:00, 680.84it/s]
64%|######3 | : 635/999 [00:00<00:00, 685.91it/s]
66%|######5 | : 659/999 [00:00<00:00, 686.73it/s]
68%|######7 | : 679/999 [00:00<00:00, 681.28it/s]
70%|######9 | : 699/999 [00:00<00:00, 676.08it/s]
72%|#######2 | : 720/999 [00:01<00:00, 673.11it/s]
74%|#######4 | : 743/999 [00:01<00:00, 673.24it/s]
77%|#######6 | : 769/999 [00:01<00:00, 677.48it/s]
80%|#######9 | : 795/999 [00:01<00:00, 681.57it/s]
82%|########2 | : 824/999 [00:01<00:00, 688.59it/s]
85%|########5 | : 852/999 [00:01<00:00, 694.45it/s]
88%|########7 | : 878/999 [00:01<00:00, 697.83it/s]
91%|######### | : 905/999 [00:01<00:00, 702.29it/s]
94%|#########3| : 937/999 [00:01<00:00, 711.50it/s]
97%|#########6| : 968/999 [00:01<00:00, 719.37it/s]
99%|#########9| : 994/999 [00:01<00:00, 721.72it/s]
100%|##########| : 999/999 [00:01<00:00, 733.16it/s]
Computing cluster p-values
Done.
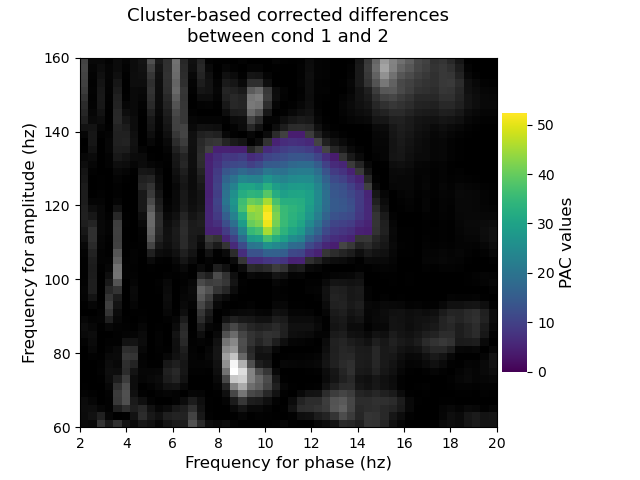
Plot the significant clusters¶
Finally, we plot the significant clusters. To this end, we used an elegant solution proposed by MNE where the non significant part appears using a gray scale colormap while significant clusters are going to be color coded.
# create new stats image with only significant clusters
t_obs_plot = np.nan * np.ones_like(t_obs)
for c, p_val in zip(clusters, cluster_p_values):
if p_val <= 0.001:
t_obs_plot[c] = t_obs[c]
t_obs[c] = np.nan
title = 'Cluster-based corrected differences\nbetween cond 1 and 2'
p.comodulogram(t_obs, cmap='gray', colorbar=False)
p.comodulogram(t_obs_plot, cmap='viridis', title=title)
plt.gca().invert_yaxis()
plt.show()
Total running time of the script: ( 0 minutes 19.566 seconds)
